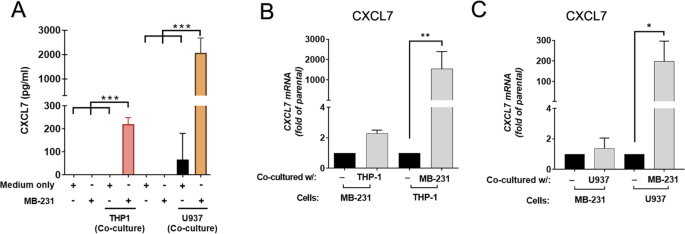
Monocytes secrete CXCL7 to promote breast cancer progression Yi-Hsiang Wang1,2 na1, Chia-Yi Shen3 na1, Sheng-Chieh Lin4, Wen-Hung Kuo5, Yuan-Ting Kuo3, Yu-Ling Hsu3, Wen-Ching Wang6,7, Kai-Ti Lin orcid.org/0000-0001-8656-57553,8 & Lu-Hai Wang orcid.org/0000-0003-0050-85602,4 Cell Death & Disease 12, Article number: 1090 (2021) Cite this article Breast cancerCancer microenvironmentChemokinesTarget identification Certain immune cells and inflammatory cytokines are essential components in the tumor microenvironment to promote breast cancer progression. To identify key immune players in the tumor microenvironment, we applied highly invasive MDA-MB-231 breast cancer cell lines to co-culture with human monocyte THP-1 cells and identified CXCL7 by cytokine array as one of the increasingly secreted cytokines by THP-1 cells. Further investigations indicated that upon co-culturing, breast cancer cells secreted CSF1 to induce expression and release of CXCL7 from monocytes, which in turn acted on cancer cells to promote FAK activation, MMP13 expression, migration, and invasion. In a xenograft mouse model, administration of CXCL7 antibodies significantly reduced abundance of M2 macrophages in tumor microenvironment, as well as decreased tumor growth and distant metastasis. Clinical investigation further suggested that high CXCL7 expression is correlated with breast cancer progression and poor overall survival of patients. Overall, our study unveils an important immune cytokine, CXCL7, which is secreted by tumor infiltrating monocytes, to stimulate cancer cell migration, invasion, and metastasis, contributing to the promotion of breast cancer progression. Breast cancer is the most commonly occurring cancer and the leading cause of cancer-related death in women worldwide [1]. Due to metastasis and resistance to systemic therapy, the mortality of breast cancer accounts for 15.5% of cancer-related death in females [1]. In recent years, the landscape of tumor microenvironment (TME), especially tumor infiltrating immune cells and cytokines, has been recognized as key factors in affecting cancer progression [2]. Targeting TME, particularly for molecules related to breast cancer metastasis, should be considered for future development of breast cancer therapy.Macrophages are important immune components in all tissues, where they play important roles in innate immunity [3]. Among the various immune cells present in the TME, tumor-associated macrophages (TAMs) are the predominant population of tumor-infiltrating immune cells [4]. Presence of TAMs in breast cancer is associated with poor prognosis and correlates with drug resistance [5]. The TAMs in breast cancer are largely derived from bone marrow monocytes that are recruited to the TME through inflammatory cytokines released by cancer cells [6]. After arriving at the TME, monocytes differentiate into macrophages by colony-stimulating factor 1 (CSF1, also known as M-CSF) [7]. In breast cancer, most TAMs will be further polarized into immunosuppressive M2-like macrophages [5] by cytokines such as interleukin (IL)−4, IL-10, and IL-13 [8]. In TME, TAMs promote cancer progression by participating in the tumor growth, angiogenesis, cell invasion, cell survival, and immune suppression [4]. Specifically, TAMs secrete matrix metalloproteinases (MMPs), serine proteases, and cathepsins to disrupt cell–cell junctions facilitating cancer cells intravasation and extravasation [9]. On the other hand, TAMs mediate immunosuppression through either expressing high levels of T-cell immune checkpoint ligands, such as PDL1, PDL2, CD80, and CD86, to directly inhibit T cells, or releasing cytokines which contribute to the maintenance of immunosuppressive TME [10, 11].Chemokines are important mediators for immune cell trafficking and differentiation [12]. In the TME, different immune cells are recruited and further differentiated via interactions between chemokines and chemokine receptors [13]. Among them, Chemokine (C–X–C motif) ligand 7 (CXCL7; also known as NAP-2), which belongs to CXC chemokine family, is originally identified as a crucial player in neutrophil recruitment upon vascular injury by binding to the CXCR2 receptor [14]. In cancer cells, CXCL7 exerts promotion of cancer development in a variety of different cancer types, including renal cell carcinoma [15], lung cancer [16], and breast cancer [17, 18], although the underlying mechanism, especially its role in metastasis, remains unclear.Here we identified a novel crosstalk between monocytes and breast cancer cells, in which monocytes secrete CXCL7 in response to CSF1 released from invasive breast cancer cells, and in turn CXCL7 acts on cancer cells. Treatment with recombinant CXCL7 protein enhanced chemotaxis of monocytes and promoted migration and invasion of breast cancer cells through FAK- and MMP13-mediated pathways. Blocking CXCL7 by a neutralizing antibody led to the suppression of tumor growth and distant metastasis in a xenograft mouse model, revealing a therapeutic potential of using CXCL7 antibody for cancer immunotherapy. Overall, our current study revealed a novel regulatory mechanism on the interplay of invasive breast cancer cells and tumor infiltrating monocytes in the TME to promote breast cancer cell growth and metastasis, and CXCL7 may serve as a potential therapeutic target for breast cancer immunotherapy.Human breast cancer cell lines (MDA-MB-231, Hs578T, BT-549) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, ThermoFisher, USA) medium containing 10% Fetal Bovine Serum (FBS, Biological Industries, USA) and 1% Penicillin/Streptomycin (P/S). Mouse breast cancer cell line 4T1 and human monocytic leukemia cell lines (THP-1, U937) were cultured in RPMI1640 (ThermoFisher, USA) medium with 10% FBS and 1% P/S. All cells were purchased from Bioresource and collection and research center (BBRC, Taiwan) or American Type Culture Collection (ATCC, USA) and authenticated by STR before shipping. MDA-MB-231 and IV2 were authenticated by STR before xenograft experiments. Mycoplasma contamination was tested if concerned. Primary bone marrow-derived mouse monocytes were from C57BL/6 mice and isolated by EasySep mouse monocytes isolation kit (Stem Cell Technologies, USA). Mouse monocytes were cultured in RPMI1640 medium containing 10% FBS and 1% P/S. All cells were cultured at 37 °C and 5% CO2 with humidity.Co-culture system and cytokine arrayCo-culture was performed using transwell chamber system (0.4-μm pores, BD Falcon, USA) in 24-well dish, in which 1 × 105 human or mouse breast cancer cells were seeded in the bottom well while 1 × 105 THP-1, U937, or mouse monocytes were seeded in the upper chamber. After 48 h incubation, the media was harvested and centrifuged at 1200 rpm to remove the cell pellet. Human Chemokine Array (C1; RayBiotech, USA) was then applied to detect differential chemokine expressions. The chemokine array experiments were carried out in duplicate.Enzyme-linked immunosorbent assay (ELISA assay)The concentration of human CXCL7 cytokine from the co-cultured media was determined by ELISA kit (RayBiotech, USA). The human CSF1 cytokine was measured using solid-phase sandwich ELISA kit (Invitrogen, USA). All procedures followed the manufacturer’s instruction. Data shown represent the means ± SD (n = 3 biological replicates).Transwell migration and invasion assaysCell migration was assayed in Falcon Cell Transwell Inserts (8.0-μm pores, BD Falcon, USA), and for the cell invasion assay, the biocoat matrigel invasion chamber was used (BD Falcon, USA) and performed as previously described [19]. Briefly, 2.5 × 104 MDA-MB-231 or Hs578T cells pre-treated with CXCL7 or pre-co-cultured with THP-1 or U937 cells were suspended in DMEM (300 μL) and seeded on the uncoated upper transwell membrane for migration assay or on matrigel-coated membrane for invasion assay. The bottom well was filled with 500 μL DMEM with 10% FBS. After incubation for 8 h (migration assay) or 18 h (invasion assay), cells on the upper side of the inserts were removed by cotton swabs, and cells adhered on the underside were fixed and stained with crystal violet. Photos of three regions were taken and the numbers of cells were counted using Image J (NIH, US). Data shown represent the normalized means ± SD (n = 3 biological replicates).Chemotaxis assayA transwell Falcon Cell Transwell Inserts (8.0-μm pores, BD Falcon, USA) was applied for chemotaxis assay. Briefly, 5 × 105 THP-1 cells were seeded on the upper chamber membrane with 8 μm pores in 24-well. The serum-free media containing different concentrations of recombinant human CXCL7 (10, 20, and 30 ng/ml) were applied in lower chamber for chemotaxis assay. Medium with 10% FBS in lower chamber was considered as the positive control. After 16 h, inserts were removed and cells migrated to the underside of the insert were collected and labeled with 1 μM CellTracker™ Red CMTPX (Invitrogen) at 37 °C for 20 min. Fluorescence intensity was measured at Ex/Em = 540/600 nm in Synergy HTX Multi-Mode Reader (Biotek, USA). The number of cells migrated toward 10% FBS was considered as 100 percent migration. Data shown represent the percentage of migrated cells ± SD (n = 3 biological replicates).Transfection & reagentsBreast cancer cells were transfected with siRNA (50 nM) by TransIT X2 (Mirus bio, USA). The siRNA sequence is listed in Supplemental Table S1. All the recombinant chemokines and antibodies used in this study were listed in Supplemental Table S2.Quantitative RT-PCR analysisRNAs were extracted from control or treated cells using TRIzol (Invitrogen) following protocols supplied by the manufacturer. First-strand cDNA was generated by ReverTraAce (Toyobo, Japan) using oligo-dT as the primer. Real-time RT-PCR was performed on qTOWER 3 Real-Time PCR Thermal Cyclers (Analytik Jena, Germany). The KAPA SYBR FAST Universal qPCR Kit (KAPA Biosystems, USA) was used. The mRNA levels were normalized to that of actin. All primer sequences used in this study are provided in Supplemental Table S3. Data shown represent the normalized means ± SD (n = 3 biological replicates).Western blot analysisCells were lysed in a 1X RIPA buffer (50 mM Tris buffer, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% SDS, and protease inhibitor mixture (Roche, USA)). The cell lysates were resolved in a 7.5–10% SDS-polyacrylamide gel, transferred onto PVDF membrane (Millipore). After protein transfer, membranes were blocked in 3% BSA-PBST (1x PBS with 0.2% Tween20) buffer at RT 30 min and then probed with indicated primary antibodies. Membranes were washed twice with PBST, then incubated with HRP-linked secondary antibody for 1 h at room temperature. After washes with PBST, ECL reagent (Millipore, USA) was used to capture luminescence by Lumin4000 (GE, USA). All antibodies used are listed in Supplemental Table S2. All experiments were repeated three times, and data shown represent the normalized means ± SE (n = 3 biological replicates).In vivo xenograft mouse model studiesFor CXCL7 antibody treatment analysis, the female SCID mice were randomly assigned to each group after arriving and injected with 1 × 106 MDA-MB-231-IV2 cells in 100 µl PBS containing matrigel per mouse at the 4th mammary fat pad at age of 4 to 8 weeks. No sample size calculation was performed. The sample size was inferred from previous studies. For intra-tumor antibody injection, each mouse tumor was injected with 20 µl of CXCL7 antibody (20 µg) or IgG1 control (20 µg) per week. For intra-venous antibody injection, tail vein was injected with 100 µl CXCL7 antibody (50 µg) or IgG1 control (50 µg) in PBS twice per week. Mice were measured for body weight and tumor size per week. After 28 days post injection, mice were sacrificed, tumors and organs (lung, axillary lymphoid node, thigh bone marrow) were harvested for RNA extraction to detect metastatic cells. No blinding was done. Relative amounts of metastatic IV2 cells from individual organ were measured by quantitative RT-PCR with human specific GAPDH levels and then normalized with actin control, which is compatible to detect actin mRNAs from both human and mouse. All procedures used in our mouse study were performed according to the approved protocol by the Institutional Animal Care and Use Committee of China Medical University, Taiwan (CMUIACUC-2018-138).Immunohistochemistry (IHC) analysisMouse breast tumor sections were prepared by pathology core lab service (NHRI), and stained with mouse macrophage marker CD206 (eBioscience, USA) and F4/80 (Abcam, USA) antibodies.Clinical samplesFrozen breast tumor samples and adjacent nontumor tissues were obtained from the Department of Surgery, National Taiwan University Hospital (NTHU), under an approved IRB protocol (201505011RINC). Samples were collected during debulking surgery, the identities of the patients remained anonymous, and informed consent was obtained from all subjects. For data from public RNA-seq database, the gene expression profile of CXCL7 and patients’ clinical information was obtained from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov) and Gene Expression Omnibus (GEO).Kaplan–Meier plotter database analysisKaplan–Meier Plotter (http://kmplot.com/analysis/) is an online database of published RNAseq array datasets that assesses the effect of 54,675 genes on survival. We performed a Kaplan–Meier Plotter analysis to assess the prognostic value of CXCL7 in patients with breast cancer or other cancer types. The hazard ratios (HRs) with 95% confidence intervals (CIs) and log-rank p-values were also computed.Statistical methodGraphPad Prism software was used for statistical analysis (GraphPad Software, Inc.). The results were shown with mean ± SD or ± SEM from three biological replicates. Student’s t test two-tail statistics analysis was used to compare two means. ANOVA followed by Tukey’s post hoc test was used for the statistical analysis when more than two means were compared. The significant p value was showed in star(s). (*, p <
https://www.nature.com/articles/s41419-021-04231-4
Monocytes secrete CXCL7 to promote breast cancer progression