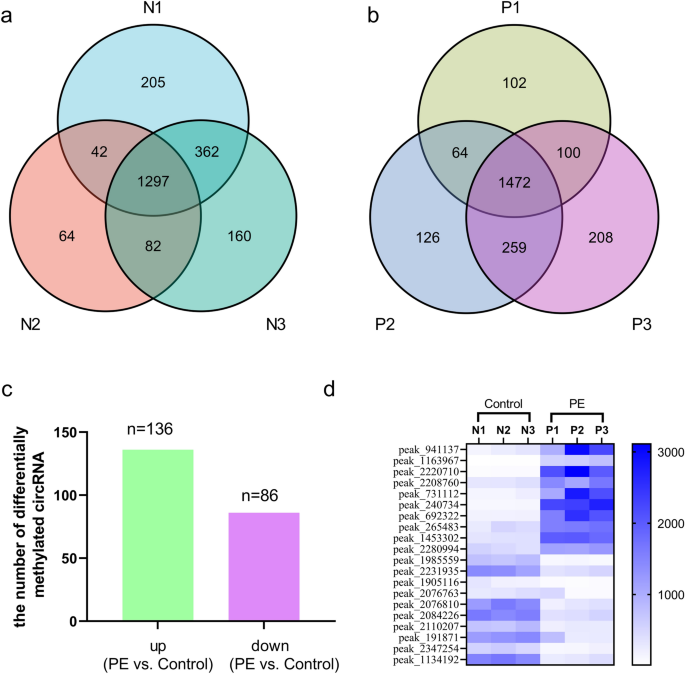
circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion Here, we performed N6-methyladenosine (m6A) RNA sequencing to determine the circRNA m6A methylation changes in the placentas during the pathogenesis of preeclampsia (PE). We verified the expression of the circRNA circPAPPA2 using quantitative reverse transcription-PCR. An invasion assay was carried out to identify the role of circPAPPA2 in the development of PE. Mechanistically, we investigated the cause of the altered m6A modification of circPAPPA2 through overexpression and knockdown cell experiments, RNA immunoprecipitation, fluorescence in situ hybridization and RNA stability experiments. We found that increases in m6A-modified circRNAs are prevalent in PE placentas and that the main changes in methylation occur in the 3’UTR and near the start codon, implicating the involvement of these changes in PE development. We also found that the levels of circPAPPA2 are decreased but that m6A modification is augmented. Furthermore, we discovered that methyltransferase‑like 14 (METTL14) increases the level of circPAPPA2 m6A methylation and that insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) maintains circPAPPA2 stability. Decreases in IGF2BP3 levels lead to declines in circPAPPA2 levels. In summary, we provide a new vision and strategy for the study of PE pathology and report that placental circRNA m6A modification appears to be an important regulatory mechanism. Preeclampsia (PE) is defined by elevated blood pressure and proteinuria or other organ damage that begins or is first recognized after 20 weeks of pregnancy1. It is one of the most common pregnancy-related disorder syndromes, with an incidence of approximately 3% to 8% worldwide2,3. PE can be injurious to the health of both the pregnant woman and the foetus. Furthermore, PE is associated with a higher and longer-term risk of cardiovascular disease development in both individuals4. Nevertheless, because the mechanism of PE pathogenesis has not yet been completely elucidated, effective means for the prevention or treatment of PE are not yet available.As the placenta is closely associated with PE, it has become a pivotal target in the study of PE pathogenic processes5. It is generally accepted in the fields of obstetrics and gynaecology that PE occurs and develops as a consequence of long-term functional dislocation and molecular abnormalities of the placenta5,6. However, the molecular pathways and precise molecular mechanisms need to be further investigated.N6-methyladenosine (m6A) is a newly identified major epigenetic modification of RNA. m6A, which was initially identified as an mRNA modification in mammals, is regarded as the most universal modification of mRNAs and non-coding RNAs in eukaryotes7,8. The latest research shows that reversible m6A modification can regulate RNA structure, stability, expression, and posttranslational modification by mediating various writer, eraser and reader proteins (such as methyltransferase‑like 3 (METTL3), fat mass- and obesity-associated protein (FTO) and YTH domain-containing family protein 2 (YTHDC2)) in all physiological and pathological situations9,10. The levels of m6A vary across different tissues11, and regulation of placental RNA m6A expression is unique.Circular RNA (circRNA) is a new kind of noncoding RNA composed of a single-stranded loop that is formed mainly from direct back-splicing of pre-mRNA. CircRNAs act as miRNA sponges, regulate target genes or proteins and encode protein functions in the course of many diseases12, indicating that circRNAs may play a key role in the progression of diseases13. Previously, we reported a large number of dysregulated circRNAs in PE placentas14. Recent studies have also suggested that circRNAs may be important in PE15.CircRNAs containing m6A modifications play important roles in many biological processes, including processes related to cancers and immune diseases16. Many studies have also shown that m6A is closely related to cardiovascular diseases, including hypertension17. Despite the publication of several articles on the relationship between m6A and hypertension disorders in pregnancy (including PE)18,19, there have been no reports on the correlation between circRNA m6A and PE. In this study, we examined the relationship between PE and circRNA m6A for the first time and sought to elucidate the mechanism of circPAPPA2 m6A modification in PE.An authorization certificate was acquired from the local medical ethics committee (Medical Ethics Committee of Shenzhen Longhua District Central Hospital, approval number: 2018080). We obtained written consent from all participants. All procedures were performed in accordance with the guidelines set forth by the Declaration of Helsinki. PE and normal control pregnancy placenta samples were collected between February 2018 and May 2019 at Guangzhou Women and Children’s Medical Centre. Placental specimens were obtained by mixing four quadrants near the maternal side after delivery, and the samples were stored at -80 °C. PE was diagnosed by a senior professor of obstetrics and gynaecology according to the 2019 criteria of the American College of Obstetricians and Gynecologists (ACOG)1. The inclusion and exclusion criteria are shown in Supplementary Table S1.Methylated RNA immunoprecipitation (MeRIP) library preparation, RNA sequencing (RNA-Seq) and data analysisThe m6A RNA-Seq service was provided by Cloudseq Biotech, Inc. (Shanghai, China). Briefly, m6A RNA immunoprecipitation (IP) was performed with a GenSeq m6A-MeRIP Kit (GenSeq, Inc., China) following the manufacturer’s instructions. Both the input samples without IP and the m6A IP samples were used for RNA-Seq library generation with an NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, Inc., USA). The library quality was evaluated with a Bioanalyzer 2100 system (Agilent Technologies, Inc., USA). Library sequencing was performed using an Illumina NovaSeq instrument with 150-bp paired-end reads.For data analysis, paired-end reads were obtained using an Illumina NovaSeq 6000 sequencer and quality-controlled according to the Q30 value. After 3′ adaptor trimming and low-quality read removal, Cutadapt software (v1.9.3) was used. First, clean reads of the input libraries were aligned to the reference genome (UCSC HG19) with STAR software, and circRNAs were identified with DCC software based on the STAR alignment results. The clean reads of all libraries were then aligned to the reference genome with HISAT2 software (v2.0.4). Methylated sites on RNA (peaks) were identified with MACS software. Differentially methylated sites were identified by diffReps. Peaks identified by both software programs with circRNA exon overlap were identified and selected with homemade scripts. Gene Ontology (GO) and pathway enrichment analyses were separately performed for differentially methylated protein-coding genes and the corresponding genes of differentially methylated circRNAs with DAVID 6.7 software. The links and version numbers of software in Table S2.Analysis of translation potentialThe open reading frames (ORFs) of the circRNAs were identified with cORF software, and the internal ribosome entry sites (IRESs) of the circRNAs were predicted with IRESfinder software. CircRNAs that met the conditions (ORF length > 150 with an IRES and with start codon (startC) regions featuring different methylation sites) were selected. The inks and version numbers of software in Table S2.Verification of m6A levels (MeRIP-qRT-PCR)Assays were carried out with a m6A-RNA methylation quantitative detection kit. Briefly, 3 μg of anti-m6A antibody (Abcam, USA, ab208577) was combined with protein A/G magnetic beads at 4 °C overnight. The antibody-bound beads and cell lysate were then incubated in IP buffer containing ribonuclease inhibitors and protease inhibitors. The bound RNAs were separated and detected by quantitative reverse transcription (qRT)-PCR.CircRNA fluorescence in situ hybridization (FISH)A direct method for FISH was implemented based on the manufacturer’s instructions to detect circPAPPA2 in TEV1 cells. Briefly, a circPAPPA2 probe was synthesized and labelled with the fluorescent substance Cy3 (RiboBio, Guangzhou, China). After fixation with 4% paraformaldehyde and treatment with 0.5% Triton X-100, cells were cultured with hybridization solution containing no probe for prehybridization to block nonspecificity. The hybridization reaction was carried out with a hybridizing solution containing a labelled probe (sequence provided in Supplementary Table S3) and cells overnight at 37 °C. Cellular DNA was counterstained with 4ʹ,6-diamidino-2-phenylindole (DAPI).qRT-PCRTotal RNA was extracted from cells or tissues with RNAiso Plus (TaKaRa, Dalian, China). After verifying the RNA quality, the RNA was reverse-transcribed using a GS II First Strand cDNA Synthesis Kit (Geneseed, Guangzhou, China). qRT-PCR was carried out with GS qPCR SYBR Green Master Mix and an ABI ViiA7 DX PCR instrument (Applied Biosystems, USA). Expression was quantified using the 2−ΔΔCt method; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. The primers used are shown in Supplementary Table S4.Immunohistochemistry (IHC)IHC was carried out according to the manufacturer’s protocol. Antigen retrieval was performed with Tris–EDTA. Hydrogen peroxide was used as an endogenous peroxidase blocker. Placental sample slides were incubated with primary antibodies (anti-insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3) antibody: ab231472, 1:100; anti-methyltransferase‑like 14 (METTL14) antibody: ab2223090, 1:200) and then with goat anti-rabbit secondary antibodies. Images were captured using a Leica microscope. A yellowish-brown colour was scored as positive (diaminobenzidine visualization).Cell transfectionCells were obtained from the American Type Culture Collection (ATCC) (Shanghai, China) and YaJi Biological (Shanghai, China); custom siRNAs targeting circRNA were obtained from RiboBio (Guangzhou, China) and cloned into a lentiviral vector by Hanheng (Shanghai, China) (the siRNA target sequence is shown in Supplementary Table S5). Overexpressed circRNA was amplified by PCR and inserted into the lentiviral vector by Hanheng. The recombinant plasmids with specific sequences and supplementary packaging plasmids were transfected into 293 T cells with Lipofectamine 2000 transfection solution. After 48 h, the viral titres were measured, and then TEV1 cells (immortalized human extravillous trophoblast cells derived from a placenta that maintain the characteristics of extravillous trophoblasts) were infected with the virus. Finally, puromycin was used to select stable strains, and the cells with overexpression or knockdown were verified by qRT-PCR.Transwell invasion assayA Transwell pore polycarbonate membrane insert (Corning, USA) was used for an invasion assay. Forty microlitres of diluted Matrigel (BD company, USA) was added to the Transwell chamber; 100 μL of serum-free medium (1 × 105 cells per well) was then added to the upper chamber, and 600 μL of serum-free medium was added to the lower chamber. The Transwell was incubated in 5% CO2 at 37 °C for 24 h, and the cells in the lower chamber were fixed and stained.Analysis of circPAPPA2 stabilityIGF2BP3-overexpressing TEV1 cells were plated in 6-well plates and reached 60% confluence after 24 h. The cells were treated with 5 μg/mL actinomycin D or DMSO for 0 h, 36 h or 72 h. The total RNA was treated with 3 U/μg RNase R and then subjected to qRT-PCR. The primers used are provided in Supplementary Table S4. The half-life of the circRNA was determined as described in previous publications20,21.RNA IP (RIP)RIP was conducted using an EZ-Magna RIP Kit (Millipore, Germany) according to the manufacturer’s instructions. Magnetic beads labelled with an anti-m6A antibody (ab185912, Abcam), an anti-IGF2BP3 antibody (ab176685, Abcam) or anti-rabbit immunoglobulin G (IgG) were incubated at room temperature for 30 min. RIP IP buffer was added to each tube. Finally, the cell lysate supernatant was added to the magnetic bead-antibody complex, and the mixture was incubated overnight at 4 °C. The coprecipitated RNA was purified and quantified by qRT-PCR.Dual-luciferase reporter assayGene fragments of the wild-type circPAPPA2 3′-untranslated region (3’UTR) carrying wild-type m6A motifs and the mutant circPAPPA2 3’UTR carrying mutant motifs (in which m6A was replaced by T) were cloned downstream of the luciferase gene in the psiCHECK2 vector. Then, we cotransfected wild-type or mutant circPAPPA2-3’UTR psiCHECK2 reporter gene plasmids and the IGF2BP3 expression plasmid or control plasmid into HEK293T cells using Cellfectin II Reagent (Invitrogen). At 48 h after transfection, Renilla and firefly luciferase activity levels were assessed.Statistical analysisStatistical analyses were performed using SPSS 24.0 (IBM) or GraphPad Prism 8.0. Differences between two groups were analysed with Student’s t-test or the Mann–Whitney U-test. P < 0.05 was considered to indicate statistical significance. The data are presented as the mean ± SD or as the number (percentage).The characteristics of the participants are shown in Supplementary Table S6.The experimental results are presented as two primary results and one secondary result. The primary results were the results of sequencing data analysis for circRNA m6A modification in PE and the results of functional/mechanistic experiments on the role of m6A-modified circPAPPA2 in PE. The secondary result was the preliminary prediction of circRNAs with translation potential.First primary result: sequencing data analysis for circRNA m6A modification in PEm6A methylation levels of circRNAs are increased in PE placentasm6A RNA-Seq analysis with MACS software revealed that the three PE subgroups shared 1472 m6A-methylated sites of circRNAs (including different sites on the same circRNA, Fig. 1A), while the three control subgroups collectively showed 1297 m6A
https://www.nature.com/articles/s41598-021-03662-5
circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion