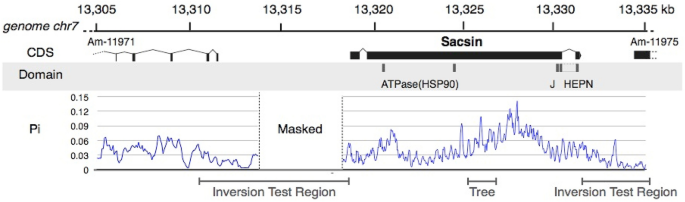
1.Hoegh-Guldberg, O. et al. Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742 (2007).ADS CAS Article Google Scholar 2.Heron, S. F., Maynard, J. A., Van Hooidonk, R. & Eakin, C. M. Warming trends and bleaching stress of the world’s coral reefs 1985–2012. Sci. Rep. 6, 38402 (2016).ADS CAS Article Google Scholar 3.Wallace, C. Staghorn Corals of the World: A Revision of the Genus Acropora (CSIRO Publishing, 1999).Book Google Scholar 4.Veron, J. E. N. Corals of the World (Australian Institute of Marine Science, 2000). Google Scholar 5.Loya, Y. et al. Coral bleaching: The winners and the losers. Ecol. Lett. 4, 122–131 (2001).Article Google Scholar 6.Feder, M. E. & Hofmann, G. E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282 (1999).CAS Article Google Scholar 7.Engert, J. C. et al. ARSACS, a spastic ataxia common in northeastern Quebec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat. Genet. 24, 120–125 (2000).CAS Article Google Scholar 8.Parfitt, D. A. et al. The ataxia protein sacsin is a functional co-chaperone that protects against polyglutamine-expanded ataxin-1. Hum. Mol. Genet. 18, 1556–1565 (2009).CAS Article Google Scholar 9.Kelley, W. L. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23, 222–227 (1998).CAS Article Google Scholar 10.Mayfield, A. B., Chen, Y.-J., Lu, C.-Y. & Chen, C.-S. The proteomic response of the reef coral Pocillopora acuta to experimentally elevated temperatures. PLoS ONE 13, e0192001 (2018).Article Google Scholar 11.Hemond, E. M., Kaluziak, S. T. & Vollmer, S. V. The genetics of colony form and function in Caribbean Acropora corals. BMC Genom. 15, 1133 (2014).Article Google Scholar 12.Weiss, Y. et al. The acute transcriptional response of the coral Acropora millepora to immune challenge: Expression of GiMAP/IAN genes links the innate immune responses of corals with those of mammals and plants. BMC Genom. 14, 1–13 (2013).Article Google Scholar 13.Fuller, Z. L. et al. Population genetics of the coral Acropora millepora: Toward genomic prediction of bleaching. Science 369 (2020).14.Shinzato, C. et al. Eighteen coral genomes reveal the evolutionary origin of Acropora strategies to accommodate environmental changes. Mol. Biol. Evol. (2020).15.Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 21, 951–960 (2005).Article Google Scholar 16.Shinzato, C., Mungpakdee, S., Arakaki, N. & Satoh, N. Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Sci. Rep. 5, 1–8 (2015). Google Scholar 17.Librado, P. & Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).CAS Article Google Scholar 18.Takahashi-Kariyazono, S., Sakai, K. & Terai, Y. Presence–absence polymorphisms of highly expressed FP sequences contribute to fluorescent polymorphisms in Acropora digitifera. Genome Biol. Evol. (2018).19.Takahashi-Kariyazono, S., Sakai, K. & Terai, Y. Presence–absence polymorphisms of single-copy genes in the stony coral Acropora digitifera. BMC Genom. 21, 1–13 (2020).Article Google Scholar 20.Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).CAS Article Google Scholar 21.Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Boil. Evol. 33, 1870–1874 (2016).CAS Article Google Scholar 22.Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+ C-content biases. Mol. Biol. Evol. 9, 678–687 (1992).CAS PubMed Google Scholar 23.Lamichhaney, S. et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat. Genet. 48, 84–88 (2016).CAS Article Google Scholar 24.Joron, M. et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477, 203–206 (2011).ADS CAS Article Google Scholar 25.Ménade, M. et al. Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations. J. Biol. Chem. 293, 12832–12842 (2018).Article Google Scholar 26.Martin, J. E., Amiot, R., Lécuyer, C. & Benton, M. J. Sea surface temperature contributes to marine crocodylomorph evolution. Nat. Commun. 5, 1–7 (2014). Google Scholar 27.Cramwinckel, M. J. et al. Synchronous tropical and polar temperature evolution in the Eocene. Nature 559, 382–386 (2018).ADS CAS Article Google Scholar Page 2Gene structure and nucleotide diversity (Pi) of the sacsin-like gene. The predicted CDS structure of the sacsin-like gene on chromosome 7 is shown, as well as the flanking downstream gene region. The positions of conserved domains in the sacsin-like gene are shown under the CDS structure. Values of nucleotide diversity estimated in sliding windows of 100 bp with a step-size of 25 bp are shown. The bar labeled ‘Inversion Test Region’ indicates the region where the PCR was performed for the inversion test. The bar ‘Tree’ indicates the region used for the construction of the phylogenetic tree.
https://www.nature.com/articles/s41598-021-02386-w
Two divergent haplogroups of a sacsin-like gene in Acropora corals