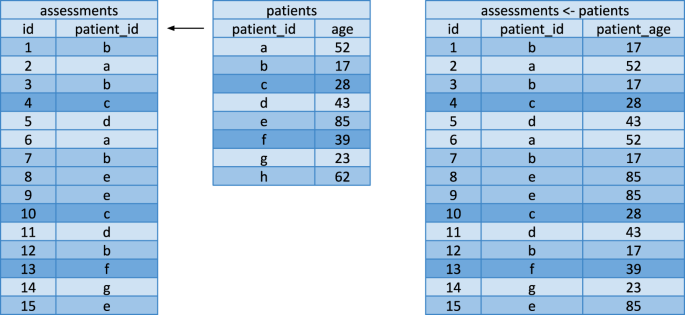
Accessible data curation and analytics for international-scale citizen science datasets Benjamin Murray orcid.org/0000-0002-2413-923X1, Eric Kerfoot1, Liyuan Chen1, Jie Deng1, Mark S. Graham orcid.org/0000-0002-4170-10951, Carole H. Sudre1,2,3, Erika Molteni orcid.org/0000-0001-7773-81401, Liane S. Canas orcid.org/0000-0002-2553-12841, Michela Antonelli1, Kerstin Klaser1, Alessia Visconti orcid.org/0000-0003-4144-20194, Alexander Hammers orcid.org/0000-0001-9530-48481, Andrew T. Chan orcid.org/0000-0001-7284-67675, Paul W. Franks6, Richard Davies orcid.org/0000-0003-2050-39947, Jonathan Wolf orcid.org/0000-0002-0530-22577, Tim D. Spector orcid.org/0000-0002-9795-03654, Claire J. Steves4, Marc Modat1 na1 & Sebastien Ourselin1 na1 Scientific Data 8, Article number: 297 (2021) Cite this article EpidemiologyResearch data The Covid Symptom Study, a smartphone-based surveillance study on COVID-19 symptoms in the population, is an exemplar of big data citizen science. As of May 23rd, 2021, over 5 million participants have collectively logged over 360 million self-assessment reports since its introduction in March 2020. The success of the Covid Symptom Study creates significant technical challenges around effective data curation. The primary issue is scale. The size of the dataset means that it can no longer be readily processed using standard Python-based data analytics software such as Pandas on commodity hardware. Alternative technologies exist but carry a higher technical complexity and are less accessible to many researchers. We present ExeTera, a Python-based open source software package designed to provide Pandas-like data analytics on datasets that approach terabyte scales. We present its design and capabilities, and show how it is a critical component of a data curation pipeline that enables reproducible research across an international research group for the Covid Symptom Study. Mobile applications have enabled citizen science1,2,3,4 projects that can collect data from millions of individuals. The Covid Symptom Study5, a smartphone-based surveillance study on self-reported COVID-19 symptoms started in March 2020, is an exemplar of citizen science. As of May 23rd, 2021, the study contains over 360 million self-assessments collected from more than 5 million individuals. The data is provided as daily CSV (comma separated value) snapshots that are made available to both academic and non-academic researchers to facilitate COVID-19 research by the wider community.The Covid Symptom Study dataset presents some demanding data curation challenges. We define data curation as involving, but not being limited to, the application of a set of transformations to the raw data. Such transformations include generation or application of metadata, cleaning of noisy and inconsistent values or relationships between values, and generation of consistent derived measures more suitable for further analysis. Erroneous values, changing schemas and multiple contemporary mobile app versions all add complexity to the task of cleaning and consolidating datasets for downstream analysis. Data curation must be performed effectively as a precondition for reproducible science. In terms of the data curation definition provided by Lee et al.6, we focus primarily on’managing and sharing data’ as defined in Table 1 of their publication.Table 1 Time taken to import the Patient table from the Covid Symptom Study 2021/05/23 snapshot.The primary challenge in curating and analysing Covid Symptom Study data is scale. Scale adds complexity to otherwise simple operations. Python-based scientific computing libraries such as Numpy7,8 and Pandas9 are ubiquitous in the academic community, but they are not designed to scale to datasets larger than the amount of RAM (random access memory) on a given machine. Commodity hardware in 2021 is typically equipped with 16 to 32 GB (gigabytes) of memory. Larger amounts of RAM necessitate expensive server-grade hardware, and doubling memory only doubles the size of dataset that can be handled.Datasets too large for standard Python scientific computing tools can be moved to datastores, either traditional, relational SQL databases or distributed NoSQL datastores, such as key-value stores. Each type of datastore comes with its own design philosophy and performance trade-offs10. Such datastores can operate on datasets far larger than RAM but involve considerable additional complexity11 through installation and maintenance burdens, and the need to learn new APIs and concepts. Additional computing power can be accessed through cloud computing but this brings ongoing costs, particularly for high-memory compute instances. Cloud computing also adds complexity to a solution, as cloud APIs are non-trivial to work with.Although most of the principal Python libraries for data science are not designed to work with larger-than-RAM datasets (see https://pandas.pydata.org/pandas-docs/stable/user_guide/scale.html), they provide a rich set of design choices and concepts that have proven successful with and are well understood by data scientists within the Python-using scientific community. By building on those design choices and concepts, but focusing on the provision of key implementation choices and algorithms that are critical for scaling beyond RAM, one can create highly scalable data analysis software with an API familiar to users of the Python ecosystem.Software packages with more scalable implementations of Pandas DataFrames exist. Dask (see https://dask.org) and PySpark12, for example, offer powerful graph-based execution models capable of performing very large calculations over multiple cores and machines. They also provide dataframe-equivalent implementations which appear promising. As we will demonstrate for Dask, the inability to cherry-pick columns from the data frame causes fundamental issues when dealing with billion element fields, however. Vaex13 is another alternative that provides similar functionalities at scale, but it is not fully open source as some components are enterprise access only.Scale is not the only challenge to consider. Reproducibility of analyses, especially across large research teams, is also critically important. When data cleaning and generation of analytics is done in an ad-hoc fashion, it is easy to generate subtly different derivations of a given measure, causing inconsistencies across research teams and research outputs. Furthermore, full reproducibility requires algorithms to be treated as immutable, so that the application of a particular algorithm to a particular snapshot of data guarantees identical results, hardware notwithstanding.The Covid Symptom Study is delivered as a series of timestamped snapshots. Corresponding entries can be modified between snapshots, and unless the dataset explicitly records changes to entries over time, one can only determine the difference by comparing the snapshots, which exacerbates scaling issues. Updating an analysis from one snapshot to another without such comparison compromises the interpretability of the updated results in a way that is less obvious when working with individual snapshots.To address the challenges described above, we have created ExeTera, a software that enables sophisticated analysis of tabular datasets approaching terabyte scale, such as the Covid Symptom Study dataset, on commodity hardware. ExeTera has an API designed to be familiar to users of Pandas, a ubiquitous tabular data analysis library within the Python ecosystem, allowing researchers to use their existing expertise. Pandas’ design is itself based on the data frame of R (see https://www.r-project.org/), another data analysis software popular with the scientific community.ExeTera can analyse datasets significantly beyond RAM scale by paying attention to two factors. The first is a careful selection of data representation that provides the ability to perform operations on tables without them ever being fully resident in memory. The second is the observation that certain operations become trivially scalable when the data is presented in lexical order. With an appropriate representation and the ability to sort table row order, commodity hardware with modest amounts of RAM can perform sophisticated analyses on datasets approaching or even exceeding terabyte scales.Although we present ExeTera in the context of the Covid Symptom Study, it is designed to be applicable to any dataset of related, tabular data. For the Covid Symptom Study, we have created ExeTeraCovid, a repository of scripts and notebooks that uses ExeTera to create reproducible end-to-end data curation workflows.The data curation workflow for the Covid Symptom Study has three stages. The first stage is a transformation of the data from text-based CSV to concrete data types where each row is parsed for validity given metadata describing the data types. The second stage is the application of a standardised set of cleaning and imputation algorithms that remove duplicate rows, detect and recover problematic values, and generate derived values from the source data. The third stage is the development and subsequent publishing of end-to-end scripts that, given a daily snapshot, can replicate a given analysis.In the following sections, we present ExeTera’s underlying capabilities and design, as well as its usage within the Covid Symptom Study as a case study.ExeTera’s performance has been benchmarked against a combination of artificial data and Covid Symptom Study data. We examine performance and scalability of ExeTera and its alternatives for key operations such as importing data, reading subsets of data, and performing joins between tables. We also demonstrate Exetera’s ability to generate a journalled dataset from snapshots allowing longitudinal analysis able to account for destructive changes between corresponding rows of the different snapshots. Finally, we present an example of ExeTera’s analytics capabilities.We have selected Pandas, PostgreSQL, and Dask to benchmark against ExeTera. Pandas has been selected as the baseline against which we are testing ExeTera. Dask has been selected as it is the most popular Python-based open-source library with a Pandas-like API that is explicitly designed for distribution and scale. PostgreSQL has been selected as it is the most popular fully open-source relational database and is widely used in academia.Performance is measured on an AMD Ryzen Threadripper 3960 × 24-core processor with 256 GB of memory. All key benchmarking processes are limited to 32 GB of memory where technically feasible although the construction of datasets for testing is allowed to use more than 32 GB. The data is read from and written to a 1 TB (terabyte) Corsair Force MP600, M.2 (2280) PCIe 4.0 NVMe SSD.DatasetsBenchmarking is performed on part of the Covid Symptom Study dataset and on synthetic data designed to mimic the relationship between the Patient and Assessment tables of the Covid Symptom Study dataset.Covid Symptom StudyWe use the Covid Symptom Study data snapshot from the 23rd of May 2021, unless otherwise stated. We make use of the three largest tables: Patients: This table has 202 columns and 5,081,709 rows. It contains biometrics, location, long term health status and other such values that rarely change over time. Assessments: This table has 68 columns and 361,190,557 rows. It includes current health status and symptoms, behavioural habits such as exposure to others and mask wearing, and other factors that are logged on an ongoing basis by contributors. Tests: This table has 20 columns and 6.979,801 rows. Tests are logged whenever a patient gets a Covid test and updated with the result of the test when available. Test parameters such as the test type and date are also recorded here. Artificial dataIn order to demonstrate the ability for ExeTera to scale relative to technologies that explicitly handle larger than RAM datasets, we construct simple artificial tables with increasingly large column counts. This is used to evaluate joins at row-counts beyond those of the Covid Symptom Study. The code to generate these tables is part of the ExeTeraEval repository, listed in the Code Availability section.The artificial dataset has two tables. The left table is an artificial analogue of the patient data from the Covid Symptom Study. The right table is an artificial analogue of the assessment data from the Covid Symptom Study. Each patient has 0 or more entries in the assessment table, with a mean of 10 entries per patient.Import performanceWe measure the performance of the import activities that must be carried out in order to perform analysis on the data. We define ‘import’ to mean anything that must be done to the data representation to minimise/eliminate the parsing required upon subsequent loads.For ExeTera, we use the exetera import operation that converts CSV data to ExeTera’s HDF5-based datastore format.For Pandas, reading from CSV imposes an expensive parse step every time a dataframe is loaded from drive, so we perform a single preliminary load from CSV as an import, assigning the appropriate metadata so that the columns are strongly typed, and then save the data as a HDF5 file. All subsequent operations are then benchmarked on the HDF5-based dataframe.For Dask, we perform a similar operation in which we read from CSV, assigning metadata and writing to partitioned HDF5 files. Note that Dask uses Pandas DataFrames for its partitions. As with Pandas, all subsequent operations are then benchmarked on the HDF5 data.For PostgreSQL, we import the data through the execution of a SQL script that creates the tables and then reads the data from CSV. We include the cost of setting up primary keys and foreign key constraints used to optimise subsequent operations. In the case of the Covid Symptom Study Patient data, this involves the removal of duplicate rows that are present in the Patient table. This allows the primary key constraint to be established on the id column and the foreign key constraint to be established on patient_id column for the Assessment and Te
https://www.nature.com/articles/s41597-021-01071-x
Accessible data curation and analytics for international-scale citizen science datasets