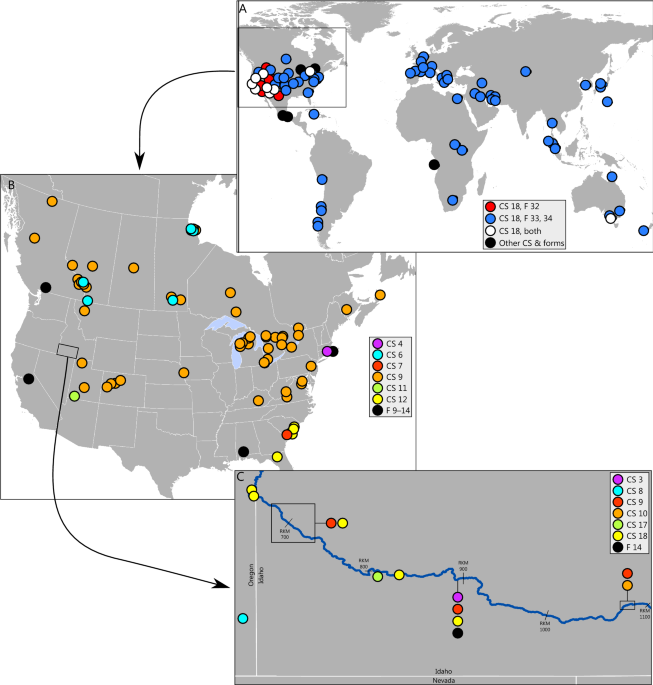
Molecular species delimitation refines the taxonomy of native and nonnative physinine snails in North America Being able to associate an organism with a scientific name is fundamental to our understanding of its conservation status, ecology, and evolutionary history. Gastropods in the subfamily Physinae have been especially troublesome to identify because morphological variation can be unrelated to interspecific differences and there have been widespread introductions of an unknown number of species, which has led to a speculative taxonomy. To resolve uncertainty about species diversity in North America, we targeted an array of single-locus species delimitation methods at publically available specimens and new specimens collected from the Snake River basin, USA to generate species hypotheses, corroborated using nuclear analyses of the newly collected specimens. A total-evidence approach delineated 18 candidate species, revealing cryptic diversity within recognized taxa and a lack of support for other named taxa. Hypotheses regarding certain local endemics were confirmed, as were widespread introductions, including of an undescribed taxon likely belonging to a separate genus in southeastern Idaho for which the closest relatives are in southeast Asia. Overall, single-locus species delimitation was an effective first step toward understanding the diversity and distribution of species in Physinae and to guiding future investigation sampling and analyses of species hypotheses. It has been argued1 that species constitute the only level of the classification of life—systematics and taxonomy—that has objective reality. Often, however, the most fundamental characteristic of an organism from a human perspective is its name1. When we delineate a species and give it a name, we facilitate communication about its relation to its environment and to other species (ecology), its patterns of survival and activity (demography and life history), and its evolutionary history and distribution (phylogenetics and phylogeography)2. We tend to focus conservation actions on named taxa3, with the tacit assumption that their members can be unambiguously tallied as present or absent, their abundance estimated and monitored, and their status as native or introduced known. Yet all species and their names are hypotheses subject to acceptance, revision, or rejection, and discerning when a name represents one species, a few populations within a species, or a complex of species is crucial to taxonomy and conservation.Robustly defining species among gastropods has been a particular challenge. Many are small and the taxonomy has generally been based on characteristics we can see, i.e., shell or soft-part morphology, which have been shown to vary in response to environmental factors or exhibit extreme conservatism among evolutionary lineages, and thus be of uncertain value for diagnosing species4,5,6,7. Consequently, species hypotheses and their higher-order assignments to genera, families, and orders could generously be described as fluid, and even authorities attempting to categorize extant species have not reached consensus, e.g., contrast8 with9. Although geography can also be informative with regard to species delineation, the recent history of many gastropods involves their widespread translocation by humans10,11 to the extent that their continental origins are sometimes uncertain12 and what were thought to be rare local endemics are instead members of globally invasive species13,14,15. At the opposite extreme, many recognized taxa are legitimately regarded as endangered because they are known from only one or a handful of sites and are restricted to freshwater habitats that are typically extensively modified for human uses16. Consequently, establishing valid species hypotheses is a critical issue.Increasingly, molecular tools are being applied to resolve taxonomic and conservation issues among taxonomic divisions within gastropods17, but this has also been unevenly applied and contentious. Although the mitochondrial genome, often the workhorse for phylogenetic efforts because of its successful application to the majority of animal taxa18, has been problematic for revealing deep phylogenetic structure in gastropods19,20,21, it has been effective for detecting relationships among genera22 and for assignment of individuals to species10, the latter the primary goal of DNA barcoding23. Mitochondrial sequences are increasingly used as a first approximation for delimiting species among taxonomically challenging groups24, but this application is rendered more difficult in analyses of gastropods because of uncertainty about what constitutes the transition from intraspecific variation to interspecific differences. For examples, some authors view combinations of highly divergent lineages (genetic distances > 15%) as constituting a single taxon7,25, whereas others6,26,27 favor thresholds for interspecific divergence akin to those applied to vertebrates (e.g., 1–4%)18,28.Gastropod snails in the subfamily Physinae (Physidae) have long posed a problem for taxonomists. Although they constitute a clade within a strongly supported, monophyletic Physidae in mitochondrial trees (Supplemental Fig. 1)9, species- and genus-level membership in Physinae remain unstable. At one time, all members were placed in the genus Physa, but subsequently have been variously assigned to Beringophysa, Haitia, Costatella, Petrophysa, Physella, Sibirenauta, Utahphysa, and Stenophysa, the latter sometimes grouped with members of the genus Aplexa in the subfamily Aplexinae8 or left unassigned29. Unsurprisingly, there are also substantial differences among different authors with respect to the identity and number of physinine species in North America9,30,31, even among scientific bodies charged with maintaining a valid taxonomy (Supplemental Table 1). In part, this may have arisen because some physinine snails are ecological generalists whose appearance is plastic in response to environmental characteristics and the presence of predators4,32. All are capable of self-fertilization, which can contribute to rapid evolution and lead to the long branches associated with Physinae in several phylogenies21. Such divergence may be accentuated in isolated or thermally enhanced habitats where founder effects are pronounced, populations are small, and generation times may be short33.Members of the physinine fauna of the Snake River basin in southern Idaho, USA are exemplars of many of these taxonomic issues. Taylor34 first described Physa (Haitia) natricina (hereafter, Physella natricina) as having a restricted distribution in a portion of the Snake River main stem, and shortly thereafter the species was listed under the US Endangered Species Act35. Authors of a subsequent morphological study of thousands of specimens from the Snake River36 argued that P. natricina did not constitute a valid taxon, and that all specimens from the Snake River were instead P. acuta, at the time thought to be introduced from Europe where it was first described in 1805. Only more recently was it recognized that the globally invasive P. acuta was actually indigenous to North America12. Regardless of its origins, its presence in the Snake River was questioned in a subsequent study of newly acquired and museum specimens from the Snake River37, whose authors countered that not only was P. natricina sufficiently morphologically and genetically discrete to merit recognition, but that the thousands of other specimens in their dataset were P. gyrina, not P. acuta. Adding to the regional complexity is a candidate species from a spring complex in Oregon in the Owyhee River basin, a tributary to the Snake River, that appears to constitute a valid taxon that has yet to be described38, and the suspected presence of an unknown number of introduced species as well as native species of dubious validity30.Initially, we planned to use molecular tools to perform specimen assignment on a sample of unidentified physinine gastropods from the Snake River basin in Idaho, USA, that were collected as part of an application for hydropower relicensing (Michael Stephenson, Idaho Power Company, personal communication). The diversity of lineages we encountered, including the presence of several potentially new species, required a broader phylogenetic scope. Hence, our objectives were to conduct molecular species delimitation among Physinae from this region and in public databases using a single mitochondrial locus and variety of approaches, to corroborate those analyses for locally obtained samples with sequences of a single nuclear gene, and to assign specimens to species using molecular tools to understand the geographical characteristics of the evolutionary lineages.Species delimitation methods were relatively consistent and often corroborated the current taxonomy but recognized greater diversity (Fig. 2, Supplemental Fig. 2). The best-scoring ASAP analysis delimited nine species, but its distance threshold (14%) was more typical of intergeneric rather than interspecific distances and tended to combine well-established and divergent taxa, e.g., one candidate species consisted of specimens of Physa fontinalis, Beringophysa jennessi, Physella pomilia, and Physella gyrina. Three of the ten top-scoring models had distance thresholds of 5.17, 5.57, and 6.28% and delineated 25, 24, and 22 species; we chose the first as the most plausible initial estimate of species diversity (see Supplemental File 1). Statistical parsimony network analyses generated 34 independent networks at the 90% threshold. Higher threshold values generated higher numbers of independent networks (e.g., 95% threshold, 39 species; data not shown) that were less consistent with the other methods and the existing taxonomy. Analyses using multi-rate Poisson tree processes delineated 22 species, albeit sometimes in combinations of specimens unsupported by the other analyses. For example, one candidate species consisted of all members of the first major clade in Physinae, despite that the maximum intraspecific distance was 23.7%. The maximum-likelihood phylogeny of histone sequences (Supplemental Fig. 3) offered less resolution among candidate taxa, but still recovered Physinae as a monophyletic clade represented by four distinct groups, with two groups representing single candidate taxa (CS 3 and 8) and two representing multiple candidate taxa (one composed of CS 9 and an undelimited taxon, the other of CS 10 and 18). Taking into account all lines of genetic, morphological (based on field identifications), and geographical evidence (Fig. 1, Supplemental Figs. 2–4, Supplemental Table 2), we delimited 18 candidate species (Table 1). Specimen assignment to a candidate species was usually straightforward; only four of the additional 232 specimens that were considered could not be assigned. Inclusion of additional specimens caused shifts in the within-tree position of clades representing candidate species, but rarely of general levels of bootstrap support for them (Supplemental Fig. 5).Figure 1Distribution of candidate species (CS) and forms (F) of members of Physinae. (A) Members of the Physella acuta sensu lato complex (CS 13–18, F 22–26). (B) Members of all other candidate species and forms in the US and Canada, excluding specimens from the Snake River basin. (C) Members of candidate species and forms found in the Snake River basin, Idaho-Oregon. The base maps were initially prepared in ArcGIS (https://www.arcgis.com/index.html) and modified in Inkscape 1.1 (https://inkscape.org).Table 1 Candidate species of Physinae delimited in this analysis; members are in Fig. 2, Supplemental Fig. 2.Figure 2Maximum-likelihood phylogeny of Physinae based on COI haplotypes and the results of species delimitation analyses. CS denotes candidate species; species labels are in Table 1 and sequence labels are in Supplemental Fig. 2. Dots (white, 85–90%; gray, 90–95%; black, > 95%) denote ultrafast bootstrap support.The geographical distribution of candidate species often did little to inform species boundaries (Fig. 1). There was widespread range overlap among candidate species, and more than one proposed taxon was often collected at a single location. Sometimes, candidate taxa were unlikely to be indigenous to the only location in which they were found (see below), implying that introductions have been widespread. This was further emphasized by the distribution of specimens in the Snake River (Fig. 1C), in which the highest diversity of candidate species was collected immediately downstream from the reach featuring a high concentration of aquaculture facilities.Below, we review these candidate species in their order of appearance in the maximum-likelihood phylogeny used in species delimitation. Monophyletic groups insufficiently diverged to constitute candidate taxa in our analyses, but recognized by some methods, were considered forms and labeled with their statistical parsimony network designation (Table 1, Supplemental Fig. 2, Supplemental Table 2) for discussion. We also note one group excluded from species delimitation—because of insufficient sequence length—that appeared in the specimen assignment phylogeny. Stenophysa and unassigned taxaThis highly supported (BS 96) clade was sister to and highly divergent from nearly all other members of Physinae. It included two specimens of Stenophysa marmorata (CS 1) from the Caribbean that constituted a robustly delimited species according to most methods. Also in this clade, however, were two specimens from southeastern Asia and a handful of specimens from one site (river kilometer 899) on the Snake River. Although these specimens group with Stenophysa marmorata, this may be a consequence of long-branch attraction. In the COI amino acid phylogeny, the unidentified specimens constitute a cohesive clade that is sister to all other members of Physinae and does not group with S. marmorata. Ng et al.14 recognized the novelty of the southeastern Asian specimens, rejected that they were introduced forms of S. spathidophallus8, and opined that they represented a new species. We take this a step farther in suggesting that the specimens from southeastern Asia and those in the Snake River constitute sister but separate candidate taxa (CS 2 and 3, respectively; minimum interspecific distance, 7.0%). Ironically, it seems possible that the specimens in the Snake River that constitute a new species are introduced, given their position immediately downstream from aquaculture facilities and their divergence from all other specimens of Physinae in North America (minimum interspecific distance, 23.4%). Moreover, this level of divergence is more consistent with their assignment to a separate genus, but whether this should be Stenophysa or a new genus requires additional samples and more comprehensive genetic analyses. A single specimen from Japan (GenBank accession LC381493, identified as Physella acuta) also grouped with CS 3 in the histone phylogeny, but could not be assigned to that species because it lacked a COI sequence and because there are no comparable histone sequences of representatives of CS 1 and 2. Physa and Beringophysa Another group of taxa forming a strongly supported (BS 96) clade consisted of one European and two North American members delimited as candidate species—Physa vernalis, P. fontinalis, and Beringophysa jennessi (CS 4–6). A fourth recognized species, P. skinneri, lacked sequences of sufficient length for species delimitation, but formed a weakly supported (BS 80) clade in the specimen assignment tree, assuming that a specimen of B. jennessi (GenBank access
https://www.nature.com/articles/s41598-021-01197-3
Molecular species delimitation refines the taxonomy of native and nonnative physinine snails in North America